Abstract
Introduction
VDJH usage description in B-cell hematological malignancies has brought new insights into the clonal differentiation and clinical implications for certain pathologies, improving patients' care, although no correlation has been made in MM.
We present the largest-to-date VDJH gene repertoire analysis in MM, consisting in biological and clinical data from 413 patients. This database was used to comprehensively investigate the characteristics of VDJH rearrangements: gene segment usage, somatic hypermutation (SHM), CDR3 characterization, and immunoglobulin stereotyped clusters assessment. We also investigated the potential relationship of these molecular features with the outcome.
Methods
Newly diagnosed MM patients were included in the study. All were managed according to the recommendations of the GEM-PETHEMA Spanish MM group, including the inclusion of most patients in the GEM2000 and GEM2005 schemes. Monoclonal assessment was carried out with FR1 VH-JH amplification, following the BIOMED-2 methods. Moreover, 113 cases were also analyzed by NGS using the LymphoTrack IGH FR1 assay (Invivoscribe Tech, San Diego, CA).
VDJH usage and mutational status were analyzed with IMGT-V-Quest. ClustalX2.0 was used to perform clustering analysis with CDR3 regions from our cohort and 1117 additional sequences obtained from IMGT/LIGM-DB corresponding to unique rearrangements from both normal and tumor human B cells. Molecular and clinical data were used to perform survival studies.
Results
The overall VDJH detection rate was 92.5% (382/413), including 376 patients with only one detectable rearrangement, five with a biallelic rearrangement, and one with a biclonal rearrangement, also detectable by flow cytometry. Thus, 388 rearrangements where detected: 362 were productive, and 26 unproductive.
Gene segments were identified in productive rearrangements. VH repertoire in MM reflected its normal counterpart, with IGHV3 being the predominant selected family, followed by IGHV4, IGHV1, IGHV2 and IGHV5. IGHD and IGHJ segment usage showed a skewed distribution, with IGHD2 and IGHD3 families accounting for 55.5% of cases, and IGHJ4 and IGHJ6 accounting for 70.8%.
SHM level could be identified in 349/362 productive sequences (mean: 9.16%, SD 3.95) with statistically significant differences between certain IGHV families, namely IGHV2 was less hypermutated than IGHV1 or IGHV4. We could also collect other rearrangement features: nucleotide addition by TdT appeared in 92.5% for N1 and 88.6% for N2, with evidence of exonuclease activity in all cases. CDR3 mean length was 16±4 aminoacids (median 15, range 6-29), with preference for Gly, Ala, Asp, Tyr (~10% each one). The R/S mutation ratio was 2.39 for CDRs versus 1.74 for FWRs, showing the high influence of antigen selection over the former. No stereotyped clusters were found in our cohort.
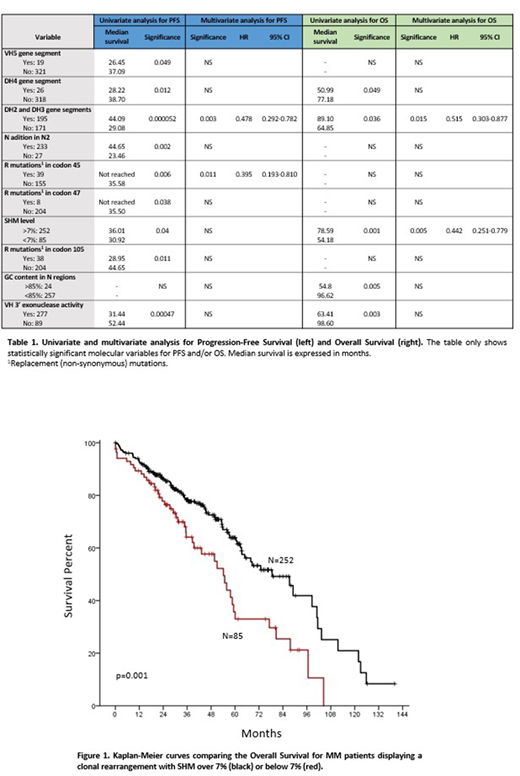
Relationship between these finding and clinical evolution was done through univariate and multivariate analyses. A shorter Progression-Free Survival (PFS) related to well-known clinical factors: presence of plasmacytomas, poor performance status or ISS stages, response to therapy, and poor-risk cytogenetics (p<0.05), as well as with gene segment selection (IGHV5, IGHD4, non IGHD2 and IGHD3), and SHM below 7%, among others (Table 1). Similar clinical data were observed for Overall Survival (OS), where the association between a short overall survival and low SHM rate (<7%) was remarkable (Figure 1).
Conclusions
VDJH usage somatic hypermutation levels and CDR3 composition in multiple myeloma resembles the normal mature B cell repertoire. In contrast to Chronic Lymphocytic Leukemia (CLL), Mantle Cell Lymphoma or Marginal Zone Lymphoma, stereotyped receptors were absent in MM, which shows no evidence of autoantigen-driven selection.
Cases with SHM levels below 7% were associated with aggressive outcome that, in the same line of CLL, would reflect the expansion of more immature clones in these cases.
More studies are needed to deepen in the clinical meaning of these new findings.
Puig:Celgene: Honoraria, Research Funding; Janssen: Consultancy, Honoraria, Research Funding; Takeda: Consultancy, Honoraria. Ocio:Takeda: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Seattle Genetics: Consultancy; Pharmamar: Consultancy; AbbVie: Consultancy; Novartis: Consultancy, Honoraria; Sanofi: Research Funding; BMS: Consultancy; Amgen: Consultancy, Honoraria, Research Funding; Mundipharma: Research Funding; Celgene: Consultancy, Honoraria, Research Funding; Array Pharmaceuticals: Research Funding. Mateos:Takeda: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Amgen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Abbvie: Consultancy, Membership on an entity's Board of Directors or advisory committees; Amgen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; GSK: Consultancy, Membership on an entity's Board of Directors or advisory committees; GSK: Consultancy, Membership on an entity's Board of Directors or advisory committees; Celgene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees. Lahuerta:Amgen: Honoraria; Janssen: Honoraria; Celgene: Honoraria. San-Miguel:Celgene: Consultancy; Sanofi: Consultancy; Janssen: Consultancy; MSD: Consultancy; Novartis: Consultancy; Takeda: Consultancy; Amgen: Consultancy; Brystol-Myers Squibb: Consultancy; Roche: Membership on an entity's Board of Directors or advisory committees. García-Sanz:Incyte: Consultancy; Amgen Inc.: Research Funding; Gilead: Research Funding; Spanish Government: Research Funding; Pharmacyclics: Research Funding; Hospira: Research Funding; BMS: Consultancy, Honoraria; Takeda: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria, Other: Travel, Accommodations, Expenses.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal